Dissecting the process of plastid division in higher plants using
functional genomics approaches
Plastids arise by division from pre-existing plastids
in the cytosol. Reminiscent of their prokaryotic ancestors, chloroplast
division occurs via binary fission and due to the inherent similarities,
bacterial cell division has been used as a paradigm for chloroplast division.
In bacteria, cell division is initiated by FtsZ, a tubulin-like GTPase, which
assembles into a cytokinetic ring (Z-ring) at midcell, resulting in the
production of two identical daughter cells and correct Z-ring placement depends
on the proteins MinC, MinD and MinE. Most plants contain nuclear genes with
high similarity to FtsZ, MinE and MinD
and the Arabidopsis homologues
AtFtsZ1-1, AtFtsZ2-1, AtMinE1, and AtMinD1 all have roles in chloroplast
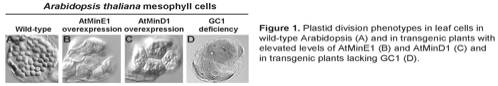
division. For example AtMinE1 (Fig. 1B) or AtMinD1 (Fig. 1C) overexpression in
transgenic Arabidopsis results in
larger and fewer chloroplasts compared to wild- type (Fig. 1A) whilst AtFtsZ
deficiency results in one chloroplast/cell.
It
is becoming increasingly clear that the different division proteins do not act
in isolation but in protein complexes. We have shown that during chloroplast
division AtFtsZ1-1 and AtFtsZ2-1 form homo and heterodimers and that AtFtsZ2-1
interacts specifically with ARC6, a protein similar to the cyanobacterial cell
division protein Ftn2. We have demonstrated that AtMinE1 forms homodimers,
acting as a topological specificity factor and that AtMinE1 interacts with
AtMinD1, which also dimerizes, and that this complex localizes to discrete
intraplastidic regions (Fig. 2A). Furthermore, we have shown that AtMinD1 is a
Ca2+-dependent ATPase and ATP hydrolysis is important for
localization and interaction with AtMinE1. Homology searches also led to the
identification of GIANT CHLOROPLAST 1 (GC1) which localizes as a dimer to the
chloroplast inner envelope. GC1 is essential in that deficiency results in
plant cells having 1-2 giant chloroplasts (Fig. 1D).
Although the Arabidopsis division machinery is evolutionary conserved division
is controlled by coordinated action of both prokaryote- and eukaryote-derived
proteins: ARC5, a dynamin-like protein and ARC3, a FtsZ-PIP5 kinase are of
eukaryotic origin and important components during plastid division. Based on our
findings to date we have constructed a working model of the molecular machinery
that controls plastid division in Arabidopsis thaliana (Fig. 2B).
The principal objective of our research is to dissect
the process of plastid division in higher plants and to determine the integral
nature of the division process within plant cells employing several functional
genomics approaches combining protein-protein interaction studies, microarray
and proteomics analysis and reverse genetics. Our sub-goals are four-fold:
1) Identify new plastid division proteins using
already characterized plastid division proteins as bait in yeast two-hybrid
screens and immunoprecipitation experiments followed by cell biological
approaches and reverse genetics to establish their role in the plastid division
process.
2) Establish the effect of different plastid division
states on nuclear gene expression dynamics using microarray analysis on plastid
division mutants with varying degrees of division defects.
3) Use comparative quantitative proteome analysis on
isolated chloroplasts from wild-type (WT) Arabidopsis and various plastid division arc mutants to determine what proteins are
recruited/de-recruited to chloroplasts and/or stabilized/destabilized within chloroplasts
during the division process. This data will complement the microarray data.
4) To understand the coordination between the stromal
plastid division machinery and the cytosolic plastid division machinery across
a double-membraned structure (Fig 3).

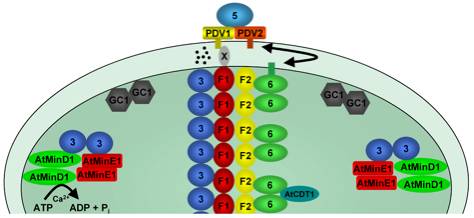
Figure 2. Model of protein-protein interactions within the
plastid division machinery. (A) Bimoecular flourescence complementation assays
have been sucessfuly used to confirm protein interations in planta. Assays were
preformed by coexpressing stromal plastid division components fused to the
N-terminal (NY) or C-terminal (CY) half of YFP and reconstituted YFP
fluorophore detected by epifluorescence microscopy. Abrevtations used: E =
AtMinE1, D = AtMinD1, F1 = AtFtsZ1-1, F2 = AtFtsZ2-1. (B) Working model for
plastid division showing the identified protein components to date, their
localization patterns and protein-protein interaction properties. AtMinE1 and
AtMinD1 localise to discrete sopts at the chloroplast membrane and interact to
form a complex. MSL2 and MSL3 (MSL) are predicted transmembrane proteins and
colocalise with AtMinE1 to the poles of the chloropalst. GC1 localizes to the
stromal side of the inner envelope membrane and forms dimers but is unable to
interact with other plastid division components. AtFtsZ1-1 (F1) and AtFtsZ2-1
(F2) form a ring-like structure at the chloropaslt mid point and can form
homodimers and heterodimers. AtFtsZ1-1 interacts with ARC3 (3) and AtFtsZ2-1
interacts with ARC6 (6). ARC3 and ARC6 both localise to ring-like structures
and both can dimerise. ARC5 localises to a ring-like structures on the
cytosolic surface of the outer envelope membrane.
Figure
3. Stromal and cytosolic plastid
division machineries. As the first step of stromal division machinery assembly AtFtsZ1-1
(F1) and AtFtsZ2-1 (F2) form a Z-ring at the centre of chloroplasts. ARC6 (6)
and ARC3 (3) are recruited to the Z-ring through specific interactions with AtFtsZ2-1
and AtFtsZ1-1, respectively. AtCDT1 also interacts with ARC6, although the
localisation of AtCDT1 is not known. The placement of the Z-ring requires the
combined action of AtMinE1, AtMinD1 and possibly ARC3, which form a complex and
can localise to plastid poles. GC1 localizes to the stromal side of the inner
envelope membrane and forms dimers. PDV1 and PDV2 localise to ring-like
structures on the cytosolic surface of the outer envelope membrane and recruit
ARC5 to the division site to constitute the cytosolic division machinery. The
inner and outer PD rings are not shown.
Coordination and signalling between the two division machineries may occur
through a direct interaction between known proteins (e.g. between ARC6 and
PDV1), may require as yet unidentified intermembrane space proteins (x) or
through the action of signalling components (black spots).
Recent publications
Jodi Maple and Simon Geir
M¿ller (2007). Plastid division
coordination across a double-membraned structure. FEBS Letters, in press
Jodi
Maple, Lea Vojta, Jurgen Soll and Simon Geir M¿ller (2007) ARC3 is a stromal Z-ring accessory
protein essential for plastid division. EMBO Rep. in press
Cassie Aldridge and Simon
Geir M¿ller (2005) The plastid
division protein AtMinD1 is a Ca2+-ATPase stimulated by AtMinE1. J. Biol.
Chem. 280. 31673-31678
Jodi
Maple and Simon Geir
M¿ller (2005).
An emerging picture of plastid division in higher plants. Planta, 223, 1-4.
Jodi
Maple, Cassie Aldridge and Simon Geir M¿ller (2005) Plastid division is
mediated by combinatorial assembly of plastid division proteins. Plant J. 43. 811-823
Jodi Maple, Cassie Aldridge and Simon Geir M¿ller (2005) The molecular biology of plastid division. J. Ex. Bot.
56, 1061-1077.
Makoto T. Fujiwara, Ayako
Nakamura, Ryuuichi Itoh, Yukihisa Shimada, Shigeo Yoshida and Simon Geir M¿ller (2004) Chloroplast division site placement requires
dimerisation of the ARC11/ AtMinD1 protein in Arabidopsis. J. Cell Science, 117, 2399-2410.
Jodi Maple, Makoto T. Fujiwara, Nobutaka Kitahata, Tracey Lawson, Neil Baker, Shigeo Yoshida and Simon Geir M¿ller (2004) GIANT CHLOROPLAST 1 is essential for correct plastid division in Arabidopsis. Current Biology. 14, 776-781.